Welcome to scorer’s documentation!¶
We propose a C++ version of the PyCM Python package. Our implementation optimizes and extends the original library including multi-threading support and an easy-to-use interface to the main algorithm. To further improve the usage of our code, we propose also a Python wrap of the library with a full compatibility with the scikit-learn package.
Usage example¶
You can use the scorer algorithm into pure-Python modules or inside your C++ application.
C++ example¶
You can use the libraries for both c++ projects, just including the scorer.h header file, and Python applications, where the scorer class is wrapped as dictionary-like object.
If you use the c++ version PAY ATTENTION to the parallel environment (you have to open the parallel region before the run of scores computation as shown in the following example code).
#include <array>
#include <scorer.h>
int main ()
{
const int n_labels = 12;
std :: array < int, n_labels > y_true = { {2, 0, 2, 2, 0, 1, 1, 2, 2, 0, 1, 2} };
std :: array < int, n_labels > y_pred = { {0, 0, 2, 1, 0, 2, 1, 0, 2, 0, 2, 2} };
scorer score;
#ifdef _OPENMP
#pragma omp parallel shared (score)
{
#endif
score.compute_score(y_true.data(), y_pred.data(), n_labels, n_labels);
#ifdef _OPENMP
}
#endif
score.print();
return 0;
}
Python example¶
The same code can be rewritten in Python (also with different labels dtype) as
#import numpy as np
from scorer import Scorer
y_true = ['a', 'b', 'a', 'a', 'b', 'c', 'c', 'a', 'a', 'b', 'c', 'a']
# np.array([2, 0, 2, 2, 0, 1, 1, 2, 2, 0, 1, 2], dtype=np.int32)
y_pred = ['b', 'b', 'a', 'c', 'b', 'a', 'c', 'b', 'a', 'b', 'a', 'a']
# np.array([0, 0, 2, 1, 0, 2, 1, 0, 2, 0, 2, 2], dtype=np.int32)
scorer = Scorer()
scorer.evaluate(y_true, y_pred)
print(scorer)
The output is the same in both the executions and it should be something like this
$ python ./scorer/example/main.py
Classes: a, b, c
Confusion Matrix:
3.0 0.0 0.0
0.0 1.0 2.0
2.0 1.0 3.0
Class Statistics:
TP(True positive/hit) 3.000 1.000 3.000
FN(False negative/miss/type 2 error) 0.000 2.000 3.000
FP(False positive/type 1 error/false alarm) 2.000 1.000 2.000
TN(True negative/correct rejection) 7.000 8.000 4.000
POP(Population) 12.000 12.000 12.000
P(Condition positive or support) 3.000 3.000 6.000
N(Condition negative) 9.000 9.000 6.000
TOP(Test outcome positive) 5.000 2.000 5.000
TON(Test outcome negative) 7.000 10.000 7.000
TPR(Sensitivity / recall / hit rate / true positive rate) 1.000 0.333 0.500
TNR(Specificity or true negative rate) 0.778 0.889 0.667
PPV(Precision or positive predictive value) 0.600 0.500 0.600
NPV(Negative predictive value) 1.000 0.800 0.571
FNR(Miss rate or false negative rate) 0.000 0.667 0.500
FPR(Fall-out or false positive rate) 0.222 0.111 0.333
FDR(False discovery rate) 0.400 0.500 0.400
FOR(False omission rate) 0.000 0.200 0.429
ACC(Accuracy) 0.833 0.750 0.583
F1(F1 score - harmonic mean of precision and sensitivity) 0.750 0.400 0.545
F0.5(F0.5 score) 0.652 0.455 0.577
F2(F2 score) 0.882 0.357 0.517
MCC(Matthews correlation coefficient) 0.683 0.258 0.169
BM(Informedness or bookmaker informedness) 0.778 0.222 0.167
MK(Markedness) 0.600 0.300 0.171
PLR(Positive likelihood ratio) 4.500 3.000 1.500
NLR(Negative likelihood ratio) 0.000 0.750 0.750
DOR(Diagnostic odds ratio) inf 4.000 2.000
PRE(Prevalence) 0.250 0.250 0.500
G(G-measure geometric mean of precision and sensitivity) 0.775 0.408 0.548
RACC(Random accuracy) 0.104 0.042 0.208
ERR(Error rate) 0.167 0.250 0.417
RACCU(Random accuracy unbiased) 0.111 0.043 0.210
J(Jaccard index) 0.600 0.250 0.375
IS(Information score) 1.263 1.000 0.263
CEN(Confusion entropy) 0.250 0.497 0.604
MCEN(Modified confusion entropy) 0.264 0.500 0.688
AUC(Area Under the ROC curve) 0.889 0.611 0.583
dInd(Distance index) 0.222 0.676 0.601
sInd(Similarity index) 0.843 0.522 0.575
DP(Discriminant power) inf 0.332 0.166
Y(Youden index) 0.778 0.222 0.167
PLRI(Positive likelihood ratio interpretation) Poor Poor Poor
NLRI(Negative likelihood ratio interpretation) Good Negligible Negligible
DPI(Discriminant power interpretation) None Poor Poor
AUCI(AUC value interpretation) Very Good Fair Poor
GI(Gini index) 0.778 0.222 0.167
LS(Lift score) 2.400 2.000 1.200
AM(Difference between automatic and manual classification) 2.000 -1.000 -1.000
OP(Optimized precision) 0.708 0.295 0.440
IBA(Index of balanced accuracy) 0.951 0.132 0.278
GM(G-mean geometric mean of specificity and sensitivity) 0.882 0.544 0.577
Q(Yule Q - coefficient of colligation) nan 0.600 0.333
AGM(Adjusted geometric mean) 0.837 0.692 0.607
MCCI(Matthews correlation coefficient interpretation) Moderate Negligible Negligible
AGF(Adjusted F-score) 0.914 0.540 0.552
OC(Overlap coefficient) 1.000 0.500 0.600
OOC(Otsuka-Ochiai coefficient) 0.775 0.408 0.548
AUPR(Area under the PR curve) 0.800 0.417 0.550
BCD(Bray-Curtis dissimilarity) 0.083 0.042 0.042
ICSI(Individual classification success index) 0.600 -0.167 0.100
Overall Statistics:
Overall ACC 0.583
Overall RACCU 0.365
Overall RACC 0.354
Kappa 0.355
Scott PI 0.344
Gwet AC1 0.389
Bennett S 0.375
Kappa Standard Error 0.220
Kappa Unbiased 0.344
Kappa No Prevalence 0.167
Kappa 95% CI (0.7867531180381775, -0.0770757719874382)
Standard Error 0.142
95% CI (0.8622781038284302, 0.30438855290412903)
Chi-Squared 6.600
Phi-Squared 0.550
Cramer V 0.524
Response Entropy 1.483
Reference Entropy 1.500
Cross Entropy 1.594
Joint Entropy 2.459
Conditional Entropy 0.959
Mutual Information 0.524
KL Divergence 0.094
Lambda B 0.429
Lambda A 0.167
Chi-Squared DF 4.000
Overall J (1.225000023841858, 0.40833333134651184)
Hamming loss 0.417
Zero-one Loss 5.000
NIR 0.500
P-value 0.387
Overall CEN 0.464
Overall MCEN 0.519
Overall MCC 0.367
RR 4.000
CBA 0.478
AUNU 0.694
AUNP 0.667
RCI 0.349
Pearson C 0.596
F1 Micro 0.583
PPV Micro 0.583
TPR Micro 0.583
SOA1(Landis & Koch) Fair
SOA2(Fleiss) Poor
SOA3(Altman) Fair
SOA4(Cicchetti) Poor
SOA5(Cramer) Relatively Strong
SOA6(Matthews) Weak
ACC Macro 0.722
F1 Macro 0.565
TPR Macro 0.611
PPV Macro 0.567
CSI
If you are working with non-integer labels the Scorer object provides a usefull _label2numbers (encode in C++) function. Its usage is mandatory in C++ since the function signature requires only integer (int32) values, while the Python version automatically encode/decode the labels according to requirements.
In the utils folder some utility scripts are reported. The make_script.py allows to write the complete parallel version of the scorer class in c++. In this way if you add a new operator to the library you can just run this code to update the scorer class parallelization strategies (computed in dependency_graphs.py script).
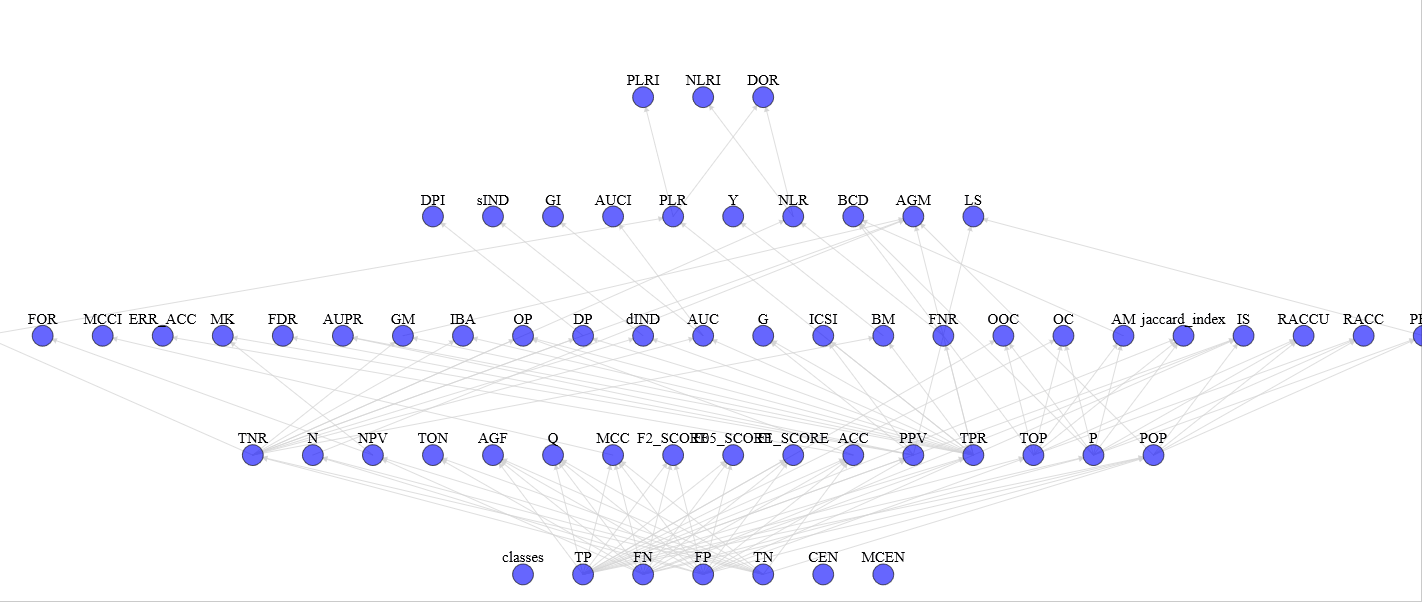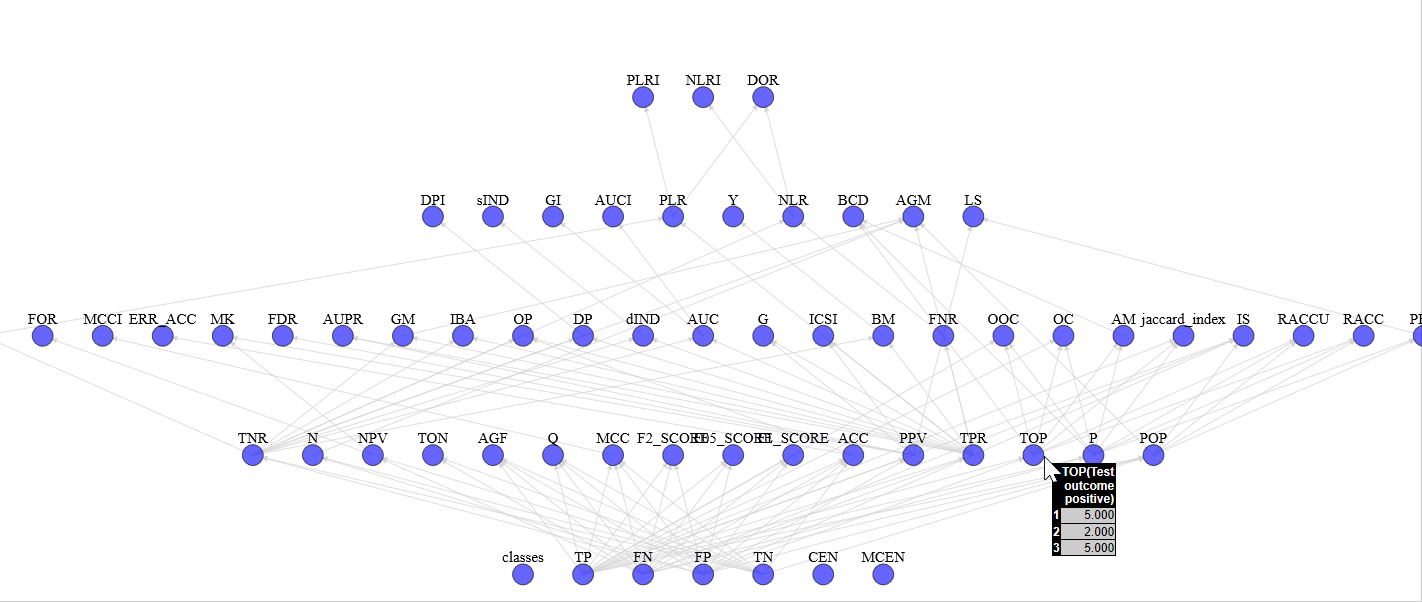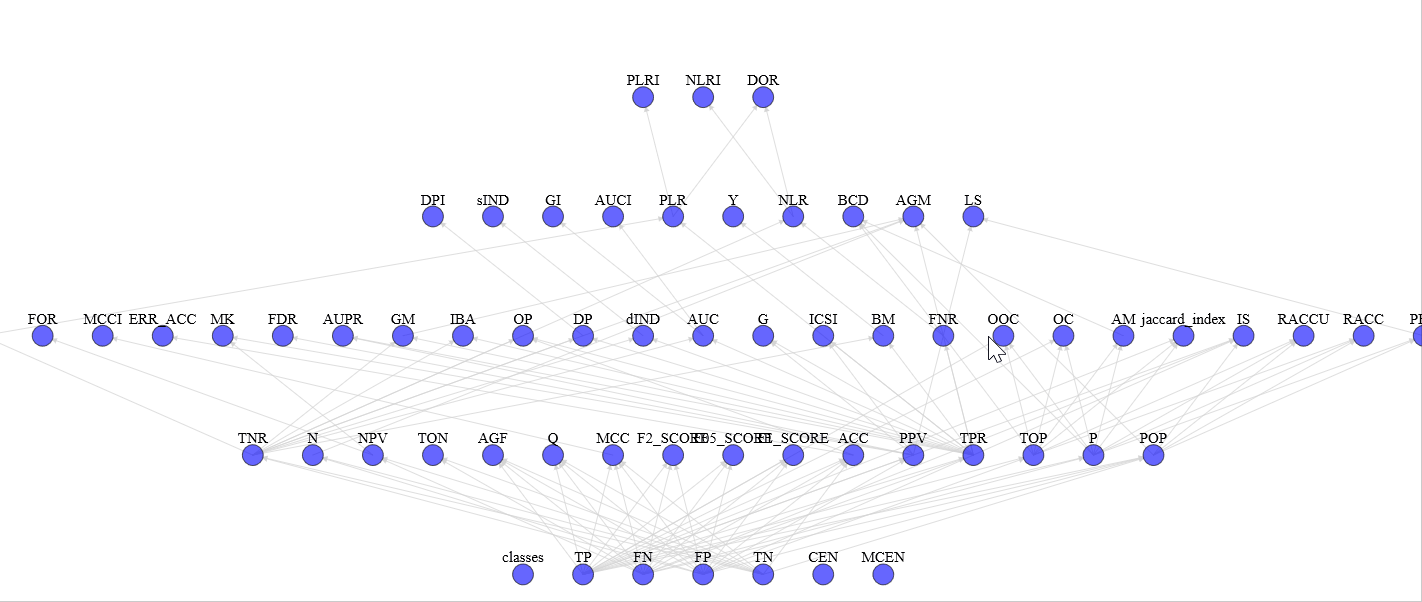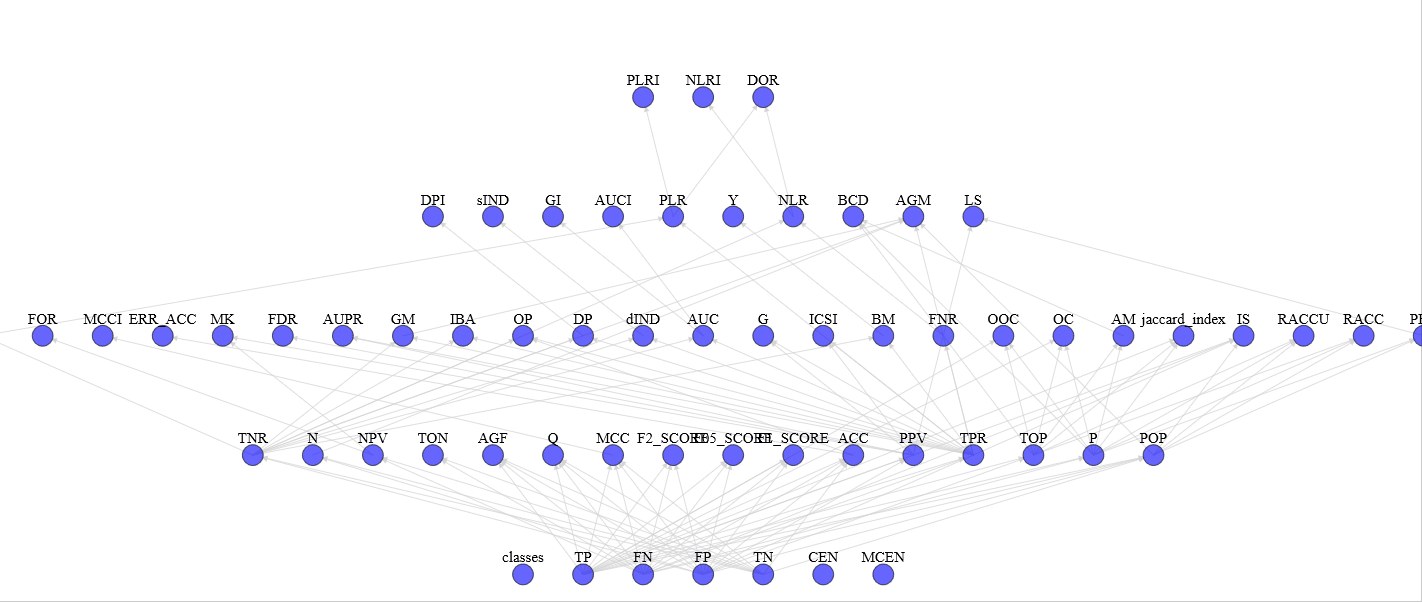
With view_stats.py you can see the complete graph of computed statistics with an HTML support for a more pleasant vision (e.g. graph)
Warning
using view_stats.py you can find the error
>>> Object of type ndarray is not JSON serializable
I suggest to use the following command to overcome this mpld3 issue:
>>> python -m pip install --user "git+https://github.com/javadba/mpld3@display_fix"